You may have heard that scientists completed the human genome (again?). What does it mean to ‘complete’ a genome? Why is this information useful, and what does this completed genome mean for science communicators? In this blog, we’ll discuss these pressing questions and the implications of the new genome.
Didn’t We Already Finish the Human Genome?
Well, yes, and no. In 2001, scientists at the National Human Genome Research Institute published the first draft of the human genome. The first draft included all the regions with accessible euchromatic chromatin, which adds up to around 92% of total genomic DNA. Using long-read sequencing technology to explore beyond that, we now have the code for the remaining 8%.
However, it’s important to note that there are as many versions of the genome as there are individual humans. So, it’s disingenuous to say we will ever be ‘finished’ sequencing the human genome, because we will realistically never sequence the genome of every single person. Instead, this latest 8% breakthrough generates a new reference genome that includes both the easily accessible euchromatic DNA and difficult to sequence highly compacted heterochromatic regions.
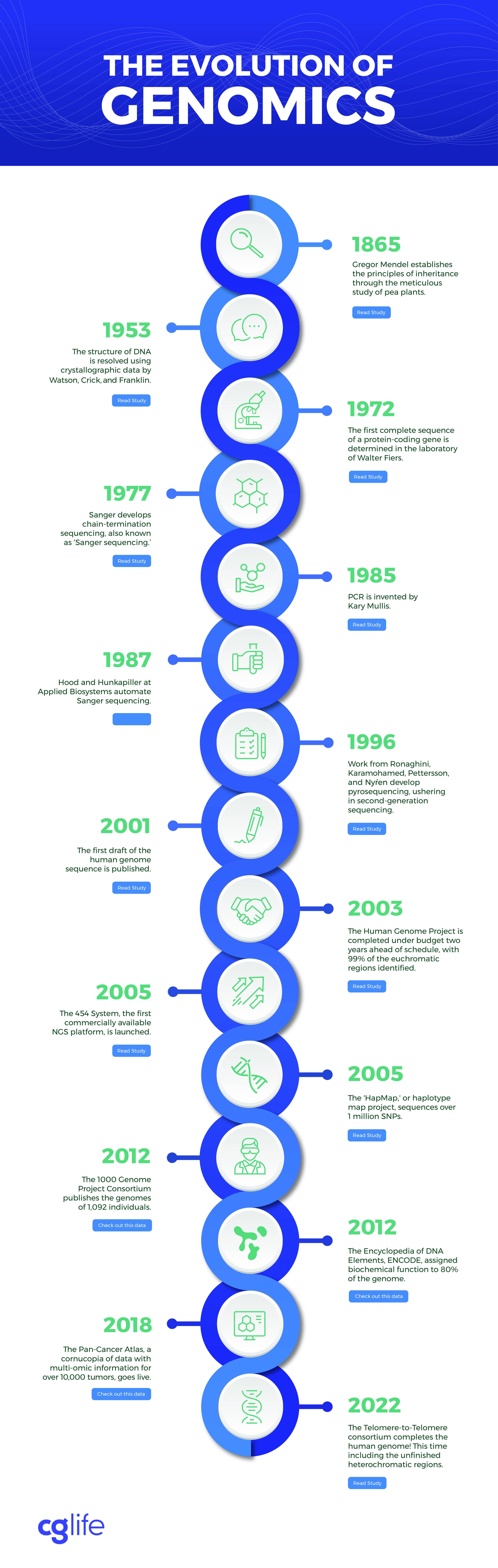 Click here to explore more with an interactive version of this timeline.
Click here to explore more with an interactive version of this timeline.
Reference Genomes are Genetic Guideposts
Whole-genome sequencing is like building a puzzle. Individual pieces start scattered randomly and are slowly assembled based on a reference. In a puzzle, your reference is the picture on the front of the box. In genomics, it’s a reference genome— our current “picture” of the human genome based on our knowledge thus far.
Because current technology can’t sequence the genome in one continuous strand, it is fractioned into millions of tiny pieces and sequenced individually. These small DNA fragments are then mapped to the most up-to-date reference, making the reference genome critically important. Without it, the pieces would be impossible to place, kind of like placing millions of puzzle pieces without any idea of the image you’re completing.
Why Did the Reference Genome Need an Update?
Completing the human genome project and thus the generation of the first reference genome was a momentous occasion. After decades of work, scientists deserved to pop champagne and celebrate this critical scientific breakthrough. However, even the authors acknowledged it was unfinished and labeled their work as an ‘essentially complete’ genome.
Scientists generated the current genome using bacterial artificial chromosomes. The genome was fractioned, inserted into bacteria for amplification, purified, sequenced, and assembled. This process doesn’t resolve repetitive, complex, and inaccessible areas in the genome well. However, complex and repetitive regions can be essential for biological function. For example, telomeres and centromeres are highly repetitive but crucial for maintaining chromosome structure. And thus, in a quest to resolve the elusive 8%, the Telomere-to-Telomere Consortium (T2T) was formed.
To overcome the challenges of the original human genome project, T2T used long-read shotgun sequencing, increasing the coverage of highly repetitive regions. Another change was the donor DNA. Where the human genome project used a series of anonymous donors for its source DNA, T2T used a single source homozygous donor DNA, making it easier to assemble the complex unidentified genomic regions.
What is Hiding in the New 8%?
We now have access to new genomic regions with documented diversity across human populations and clinical subgroups. These regions include ribosomal DNA, acrocentric chromosomes, and alpha satellite regions. Ribosomal DNA is exactly what it sounds like— the genetic code to create ribosomes. However, the coding regions for 45S rRNA are large, nearly identical repeats, making them difficult to sequence. Acrocentric chromosomes, found on chromosomes 13, 14, 15, 21, and 22, are small chromosomal arms found close to the centromeres. Interestingly, the authors found that all acrocentric chromosomes are relatively similar! Perhaps the acrocentric chromosome sequence is conserved to maintain its structure. Alpha satellite regions are highly repetitive genomic regions enriched near centromeres, which until now, have escaped complete genome assembly.
To summarize the 3.055 billion base-pair long genome published by T2T- the study unveiled an additional 200 million nucleotides with 99 predicted coding genes.
What This Means For You
The human genome project assembled the genomic puzzle at a high level, but with T2T, we now have every piece. The completed puzzle will accelerate research, particularly in previously undetermined genomic regions. Hopefully, this will bring us closer to the promise of genomics- personalized medicine, where clinical care is tailored to your genetic code.
Genomics was already moving at lightning speed. With the completed human genome, progress in genomic medicine and other applications will only get faster. Technological advances increasingly rely on genomic technologies, and it’s our role as science communicators to translate these innovations to wider audiences. As science communicators, it’s critical to develop a functional understanding of genomics and use that knowledge to increase genomic literacy broadly. We are only now beginning to understand the full picture of what makes us human genetically. I can’t wait to see how researchers will use this new information to improve human life.
